WP2: Histotyping
Histopathology refers to the microscopic examination of tissue to study the manifestation of disease. In the DoMore! project, all patients have been diagnosed with cancer, and their tumours were surgically resected.
The histopathologists study the appearance of the resected specimen and assess relevant properties, such as the tumour grade. Tumour grading is an evaluation of the extent to which tumour cells and tumour tissue resemble normal cells and tissue, where a high degree of similarity (well differentiated) is associated with a better prognosis for the patient than a low degree of similarity (poorly differentiated). The analysis is carried out in HE-stained tissue sections. Tumour grade is a good prognostic marker, but a substantial proportion of patients are classified as moderately differentiated, i.e., an intermediate group with an intermediate outcome. The increasing workload for pathologists as well as significant intra- and interobserver variability implies a need for automated methods for this task.
Histotype = Any of a range of tissue types that arise during the growth of a tumour.
Automation
We have developed Histotyping, a fully automated histological characterisation of HE-stained sections from cancer specimens for prognostic purposes. The method is based on deep learning by convolutional neural networks trained on images of HE-stained tissue sections where the patient outcome is used to guide the training process into a system that is able to identify tissue patterns in the HE-sections that are distinct for patient prognosis. The resulting computer model can be applied to a new patient’s tumour sample and estimates the probability of a poor patient outcome.
Risks and Preventions
Large amounts of labeled data are required to train this type of deep convolutional neural network and not until recent years have we had the required datasets and computational resources to perform these types of analyses. The neural network models have millions of features and overfitting to the training dataset is a common problem, i.e., that the computer model identifies and exploits artifacts in the training dataset that are associated with the desired outcome, but have no biological relevance and fails when evaluated on a new dataset on which it is not trained. To increase the probability that the neural network generalises when applied to new patients from a new dataset, we have used a robust design with thousands of patients from different patient cohorts with the same cancer type. We have developed the framework on stage I-III colorectal cancer patients from two hospitals in Norway and from two clinical trials in England. Another risk in the development of such models is the adaption to the technical equipment such as the imaging system, i.e., that the method works well on images scanned with the scanner on which it is developed and not on images from a scanner from another vendor. To compensate for this problem, we have scanned all images with scanners from two major scanner manufacturers (Hamamatsu and Leica).
A third risk in the development of a computerised system for risk assessment based on scanned HE-sections is the dependence on lab preparation, i.e., that the method works on scans of tissue sections prepared and stained in the lab where the method is developed only. To evaluate this, we have scanned parallel sections from one of the patient cohorts (Gloucester) that have been prepared in the pathology routine there.
We have partitioned our patient datasets to make the best use of the data. The Kaplan-Meier plots below illustrate results based on images scanned with 10x, and 40x lens analyzed in corresponding neural network models and classified according to the agreement between the two models’ classifications in a test partition that has not been included in the training process.
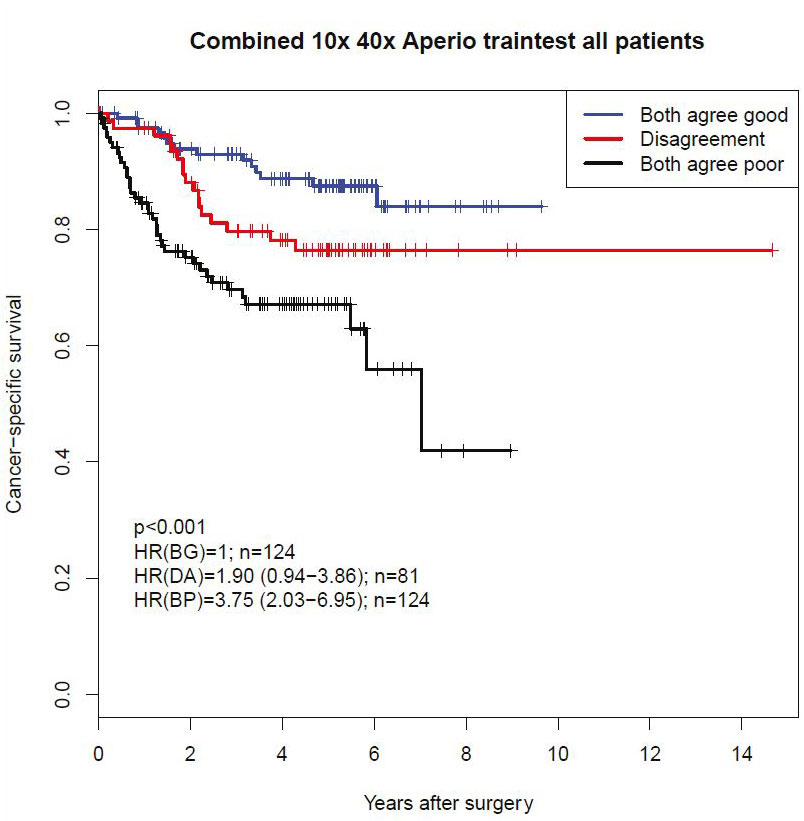 Kaplan-Meier plot of cancer-specific survival probability grouped by classification by 10x and 40x models and their agreement based on scans from the Leica Aperio scanner.
Kaplan-Meier plot of cancer-specific survival probability grouped by classification by 10x and 40x models and their agreement based on scans from the Leica Aperio scanner.
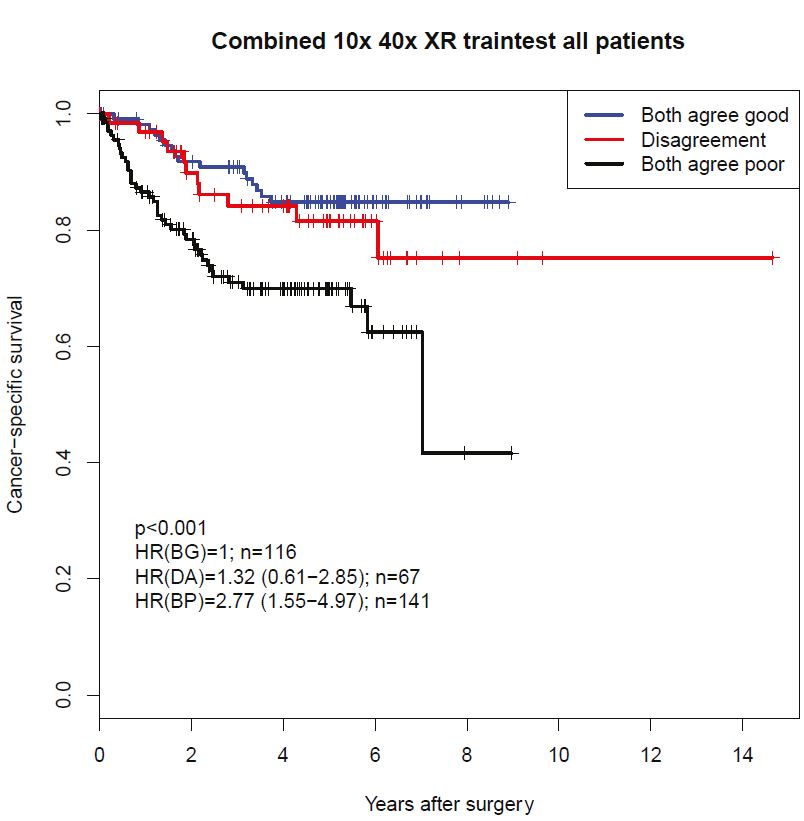 Kaplan-Meier plot of cancer-specific survival probability grouped by classification by 10x and 40x models and their agreement based on scans from the Hamamatsu XR scanner.
Kaplan-Meier plot of cancer-specific survival probability grouped by classification by 10x and 40x models and their agreement based on scans from the Hamamatsu XR scanner.
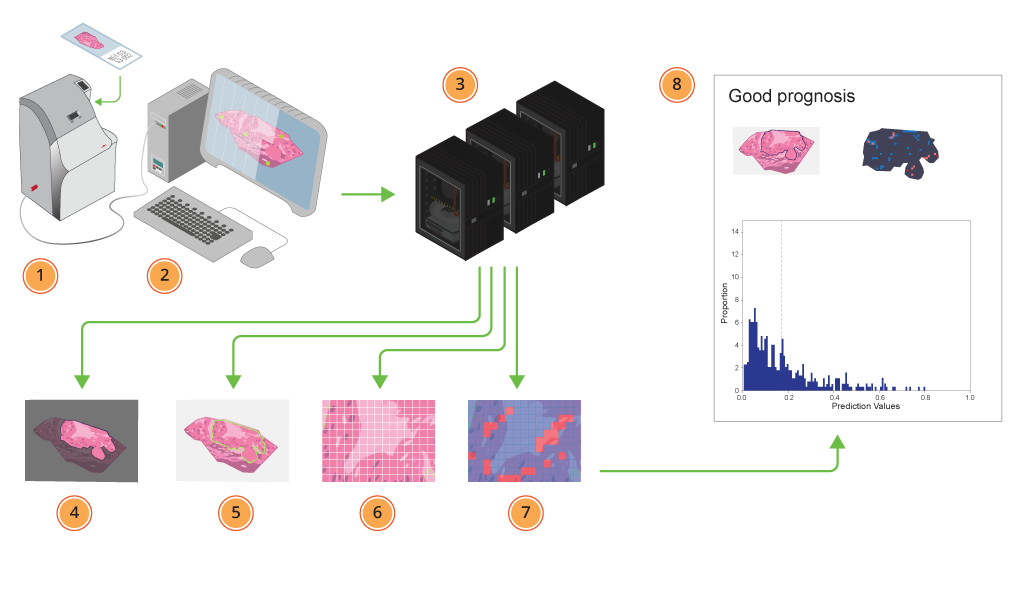 Histotyping: 1) Samples are fed into the scanner. 2) The scanner scans the tissue sections, adjusting focus to get the best result possible. 3) The scans are sent to the deep learning models, running on GPUs where 4) the tumour region in each scanned sample is automatically segmented, 5) the outlined tumour is further, 6) divided into tiles, and 7) automatically assigned a probability for representing poor prognosis. Each tile is toned blue for good, or red for a bad prognosis. 8) Finally a report is generated, where patient prognosis is estimated based on the individual tiles’ prediction values. The different tumour regions’ contribution to the patient classification are illustrated as demonstrated in the image on the left page.
Histotyping: 1) Samples are fed into the scanner. 2) The scanner scans the tissue sections, adjusting focus to get the best result possible. 3) The scans are sent to the deep learning models, running on GPUs where 4) the tumour region in each scanned sample is automatically segmented, 5) the outlined tumour is further, 6) divided into tiles, and 7) automatically assigned a probability for representing poor prognosis. Each tile is toned blue for good, or red for a bad prognosis. 8) Finally a report is generated, where patient prognosis is estimated based on the individual tiles’ prediction values. The different tumour regions’ contribution to the patient classification are illustrated as demonstrated in the image on the left page.
Research results published in the Lancet
The article "Deep learning for prediction of colorectal cancer result: a discovery and validation study" was published in The Lancet in January 2020. The institute also provided a video explaining Histotyping:
Oslo, 15th February 2020
Navigate to the other Work Package descriptions: